Source file: notebooks/umi_correction/trimmed_collisions.Rmd
Last updated: 2018-02-18
Code version: e50352c
library(ggplot2)
library(ggsci)
library(ggridges)
library(ggrastr)
library(dplyr)
library(parallel)
library(reshape2)
library(dropestr)
library(dropEstAnalysis)
theme_set(theme_base)
kPlotsFolder <- '../../output/figures/'
kDataPath <- '../../data/dropest/'
# reads_per_umi_per_cell <- holder$reads_per_umi_per_cell
# reads_per_umi_per_cell$reads_per_umi <- FilterNUmis(holder$reads_per_umi_per_cell$reads_per_umi)
reads_per_umi_per_cell <- kDataPath %>%
paste0('10x/aml035_post_transplant/est_10_20_umi_quality/reads_per_umi_per_cell.rds') %>%
readRDS()
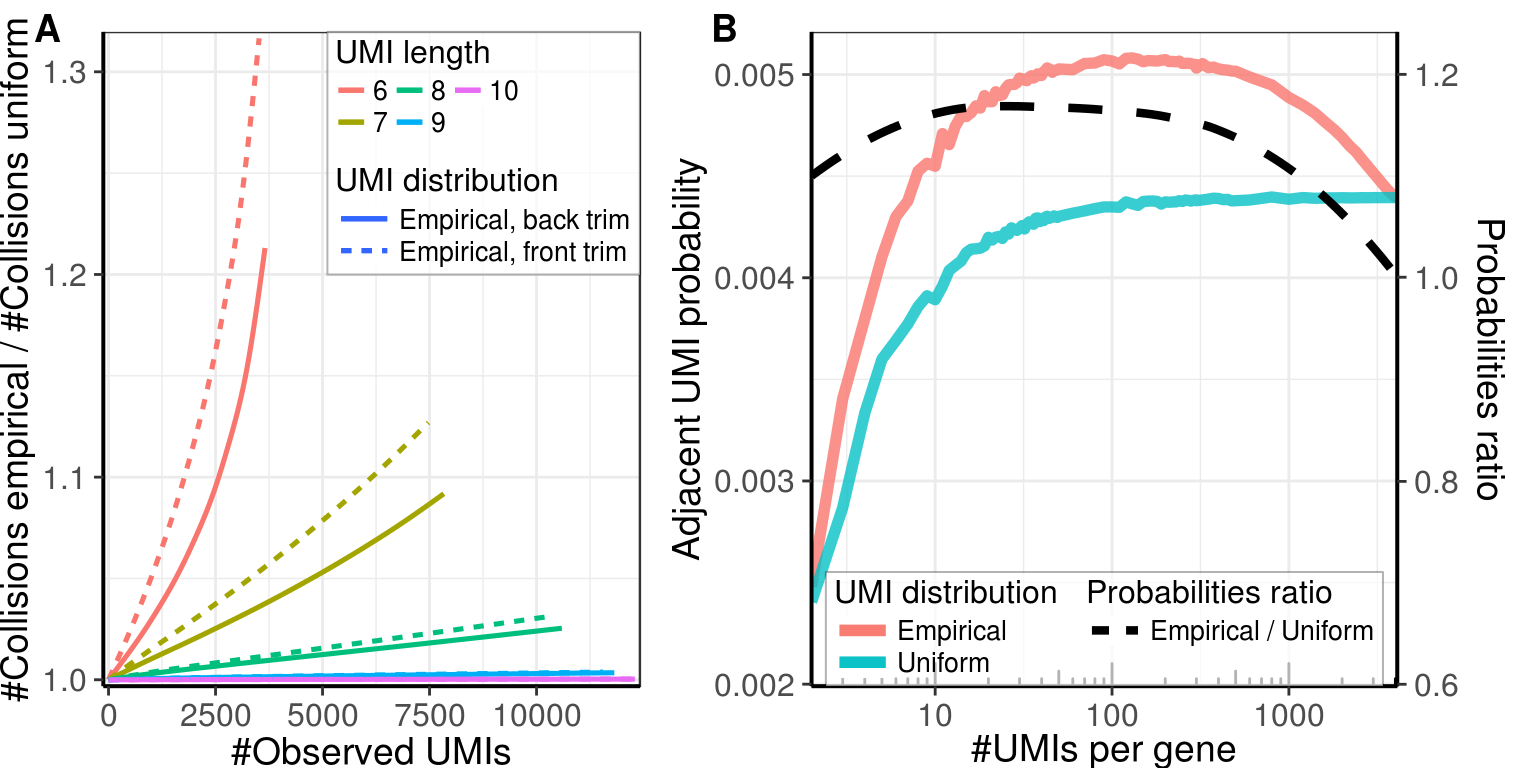
Collisions
Trimming:
umi_lengths <- rep(6:10, 2)
is_reverse <- c(rep(F, 5), rep(T, 5))
trimmed <- mcmapply(function(i, b) TrimUmisSummary(reads_per_umi_per_cell, i, b),
umi_lengths, is_reverse, SIMPLIFY = F, mc.cores=10)
collisions_infos <- lapply(trimmed, function(d) d$collisions.info)
collision_df <- mapply(function(s, l, r) data.frame(Observed=1:length(s), Adjusted=s,
UmiLength=l, IsReverse=r),
collisions_infos, umi_lengths, is_reverse, SIMPLIFY = F) %>%
bind_rows() %>%
mutate(AdjustedUniform=mapply(AdjustGeneExpressionUniform, Observed, 4^UmiLength))
collision_dfs <- split(collision_df, collision_df$IsReverse) %>%
setNames(c('Reverse', 'Direct'))
rm(trimmed, reads_per_umi_per_cell, collisions_infos, collision_df)
invisible(gc())
Main figure
plt_guide <- guides(color=guide_legend(title='UMI length', nrow = 2, order=1),
linetype=guide_legend(title='UMI distribution', order=2, keywidth = 1.5))
expand <- c(0.01, 0)
fig_collisions <- ggplot(collision_dfs$Direct, aes(x=Observed, col=as.factor(UmiLength))) +
geom_line(aes(y=Adjusted - Observed, linetype='Empirical'), size=0.9) +
geom_line(aes(y=AdjustedUniform - Observed, linetype='Uniform'), size=0.9) +
scale_x_continuous(expand = expand, breaks=seq(4000, 12000, 4000)) +
scale_y_continuous(expand = expand) +
labs(x='#Observed UMIs', y='#Collisions') +
theme_pdf(legend.pos=c(1, 1)) + plt_guide
fig_collisions <- cowplot::plot_grid(fig_collisions, labels='C')
ggsave(paste0(kPlotsFolder, '/fig_collisions.pdf'), fig_collisions, height=5.5, width=3.5)
print(fig_collisions)

rm(fig_collisions);
Ratio
gg_collisions_ext <- ggplot(mapping=aes(x=Observed, col=as.factor(UmiLength),
y=Adjusted / AdjustedUniform)) +
geom_smooth(data=collision_dfs$Direct, mapping=aes(linetype='Empirical, back trim'),
size=0.9, se=F) +
geom_smooth(data=collision_dfs$Reverse, mapping=aes(linetype='Empirical, front trim'),
size=0.9, se=F) +
scale_x_continuous(expand = expand) + scale_y_continuous(expand = expand) +
labs(x='#Observed UMIs', y='#Collisions empirical / #Collisions uniform') +
theme_pdf(legend.pos=c(1, 1)) +
plt_guide
rm(collision_dfs)
gg_collisions_ext
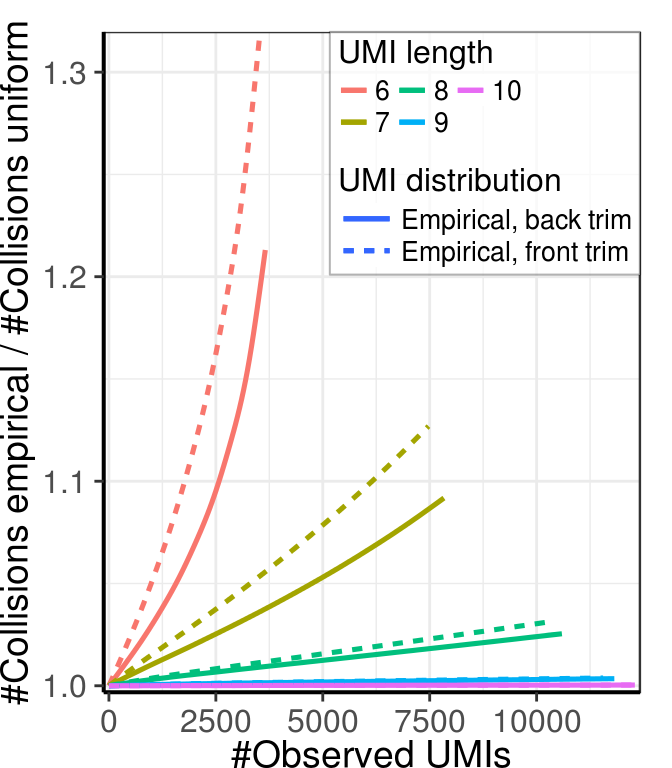
Estimated number of adjacent UMIs
holder <- readRDS(paste0(kDataPath, 'SCG71/est_01_15_umi_quality/SCG71.rds'))
umi_distribution <- GetUmisDistribution(holder$reads_per_umi_per_cell$reads_per_umi)
umi_probs <- umi_distribution / sum(umi_distribution)
rm(holder, umi_distribution)
invisible(gc())
gene_sizes <- list(2:10, 11:50, seq(60, 300, 10), seq(300, 500, 25), seq(600, 4090, 200),
c(4050, 4095))
sample_nums <- unlist(mapply(rep, c(100000, 100000, 15000, 1000, 500, 500),
sapply(gene_sizes, length)))
gene_sizes_f <- unlist(gene_sizes)
adjacent_umis_num <- mcmapply(function(s, n)
SampleNumbersOfAdjacentUmis(s, umi_probs, n, uniform=F),
gene_sizes_f, sample_nums, mc.cores=30)
adjacent_umis_num_unif <- mcmapply(function(s, n)
SampleNumbersOfAdjacentUmis(s, umi_probs, n, uniform=T),
gene_sizes_f, sample_nums, mc.cores=30)
plot_df <- data.frame(GeneSize=c(1, unlist(gene_sizes))) %>%
mutate(ProbOfAdjacentUMIs=c(0, sapply(adjacent_umis_num, mean)) / GeneSize^2,
ProbOfAdjacentUMIsUnif=c(0, sapply(adjacent_umis_num_unif, mean)) / GeneSize^2)
axis_ratio <- 200
axis_offset <- 0.001
gg_prob_ratio <- ggplot(plot_df, aes(x=GeneSize)) +
geom_line(aes(y=ProbOfAdjacentUMIs, color='Empirical'), size=2, alpha=0.8) +
geom_line(aes(y=ProbOfAdjacentUMIsUnif, color='Uniform'), size=2, alpha=0.78) +
geom_smooth(aes(y=ProbOfAdjacentUMIs / ProbOfAdjacentUMIsUnif / axis_ratio - axis_offset,
linetype='Empirical / Uniform'), size=1.5, color='black', se=FALSE) +
labs(x = '#UMIs per gene', y='Adjacent UMI probability') +
scale_x_log10(expand=c(0, 0), limits=c(2, 4100)) + annotation_logticks(side='b') +
scale_y_continuous(expand=c(1e-5, 1e-5), limits=c(0.002, 0.0052),
sec.axis=sec_axis(trans=~.*axis_ratio + axis_offset*axis_ratio,
name='Probabilities ratio')) +
scale_linetype_manual(values='dashed', name='Probabilities ratio') +
guides(color=guide_legend(title='UMI distribution')) +
guides(linetype=guide_legend(override.aes=list(linetype=6))) +
theme_pdf(legend.pos=c(0.5, 0)) +
theme(legend.box='horizontal', legend.key.width=unit(0.3, 'in'))
gg_prob_ratio
This R Markdown site was created with workflowr