Validation of filtration of low-quality cells on inDrop Mouse BMCs
Viktor Petukhov
2018-01-23
Source file: notebooks/low_quality_cells/validation_bmc.Rmd
Last updated: 2018-05-16
Code version: dae257a
library(ggplot2)
library(ggpubr)
library(ggrastr)
library(dplyr)
library(parallel)
library(Seurat)
library(dropestr)
library(dropEstAnalysis)
theme_set(theme_base)
set.seed(42)
kOutputFolder <- '../../output/'
kDataPath <- '../../data/'
kEstDataPath <- paste0(kDataPath, 'dropest/SCG71/est_11_14_poisson_real/')
kAnnotationDataPath <- paste0(kDataPath, 'annotation/')Load data
holder <- readRDS(paste0(kEstDataPath, 'SCG71.rds'))est_cell_num <- EstimateCellsNumber(holder$aligned_umis_per_cell)
umis_per_cell <- sort(holder$aligned_umis_per_cell, decreasing=T)scores <- ScorePipelineCells(holder, mit.chromosome.name='chrM', predict.all=T,
verbose=T)[names(umis_per_cell)]Explained variance after PCA: 96.78%; used 3 PCs.
Used features: ReadsPerUmi, UmiPerGene, LowExpressedGenesFrac.PlotCellScores(scores)
intersect_cbs <- names(scores[1:est_cell_num$expected])
intersect_cbs <- intersect_cbs[scores[intersect_cbs] > 0.9]
unknown_cell_scores <- scores[(est_cell_num$expected+1):length(scores)]
rescued_cbs <- names(unknown_cell_scores)[unknown_cell_scores > 0.9]
unknown_cell_scores <- scores[1:est_cell_num$expected]
filtered_cbs <- names(unknown_cell_scores)[unknown_cell_scores < 0.1]
c(Unchanged=length(intersect_cbs),
Rescued=length(rescued_cbs), Filtered=length(filtered_cbs))Unchanged Rescued Filtered
4684 126 86 r_cm_rescued <- holder$cm_raw[, c(names(umis_per_cell)[1:est_cell_num$expected],
rescued_cbs)]# You need to run "annotation/annotation_bmc.Rmd" first
clusters_annotated <- paste0(kAnnotationDataPath, 'indrop_bmc_clusters_annotated.csv') %>%
read.csv() %>% (function(x) setNames(as.character(x$Type), x$Barcode))Rescued cells
r_rescued <- GetPagoda(r_cm_rescued, n.cores=30)5558 cells, 11856 genes; normalizing ... using plain model winsorizing ... log scale ... done.
calculating variance fit ... using gam 198 overdispersed genes ... 198 persisting ... done.
running PCA using 1000 OD genes .... done
calculating distance ... pearson ...running tSNE using 30 cores:intersect_clusters <- clusters_annotated[intersect(names(clusters_annotated),
intersect_cbs)]
notannotated_cells <- setdiff(colnames(r_cm_rescued), names(clusters_annotated))
clusters_annotated_resc <- AnnotateClustersByGraph(r_rescued$graphs$PCA,
clusters_annotated, notannotated_cells,
max.iter=100, mc.cores=10)
rescued_clusters <- clusters_annotated_resc[rescued_cbs]
intersect_clusters <- clusters_annotated[intersect_cbs]plot_cbs <- names(clusters_annotated) %>% setdiff(rescued_cbs) %>% setdiff(filtered_cbs)
plot_clusters <- clusters_annotated[plot_cbs]
plot_rescued_clusters <- rescued_clusters
for (type in c("Maturing neutrophils", "Maturing macrophages", "Cycling cells")) {
plot_clusters[plot_clusters == type] <- gsub(" ", "\n", type)
plot_rescued_clusters[plot_rescued_clusters == type] <- gsub(" ", "\n", type)
}
gg_tsne <- PlotFiltrationResults(r_rescued, plot_clusters, filtered.cbs=filtered_cbs,
rescued.clusters=plot_rescued_clusters,
raster.width=4, raster.height=4.8, lineheight=0.9) +
ylim(-35, 33) +
theme_pdf(legend.pos=c(0, 1), show.ticks = F)
gg_tsne
Number of rescued cells per cluster
rescued_table <- TableOfRescuedCells(clusters_annotated_resc[c(intersect_cbs, rescued_cbs)],
rescued_cbs)
write.csv(rescued_table, paste0(kOutputFolder, "tables/rescued_cbc_bmc.csv"), row.names=F)
rescued_table| Cell type | Total num. of cells | Num. of rescued | Fraction of rescued, % |
|---|---|---|---|
| B cells, immature | 99 | 0 | 0.00 |
| B cells, mature | 457 | 1 | 0.22 |
| Cycling cells | 192 | 5 | 2.60 |
| Mast cells | 48 | 1 | 2.08 |
| Maturing macrophages | 653 | 1 | 0.15 |
| Maturing neutrophils | 2876 | 113 | 3.93 |
| NK cells | 61 | 0 | 0.00 |
| pre-B cells | 120 | 0 | 0.00 |
| Progenitors | 222 | 4 | 1.80 |
| T cells | 82 | 1 | 1.22 |
Seurat analysis
seurat_cm <- r_cm_rescued[Matrix::rowSums(r_cm_rescued) > 200, ]
srt <- CreateSeuratObject(raw.data=r_cm_rescued, project="BMC", display.progress=F)
srt <- NormalizeData(object=srt, normalization.method="LogNormalize", scale.factor=10000)
srt <- FindVariableGenes(object=srt, mean.function=ExpMean, dispersion.function=LogVMR,
x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 1, do.plot=F)
srt <- ScaleData(object = srt, vars.to.regress = "nUMI", display.progress=F)srt@ident <- as.factor(clusters_annotated_resc[colnames(srt@raw.data)])
names(srt@ident) <- colnames(srt@raw.data)
compared_clusters <- unique(srt@ident)
cluster_markers <- mclapply(compared_clusters, function(i)
mclapply(setdiff(compared_clusters, i), FindClusterMarkers, i, srt, mc.cores=4),
mc.cores=11)overexpressed_genes <- GetOverexpressedGenes(srt, compared_clusters, cluster_markers)
clusters_info <- list(clusters=srt@ident, marks=cluster_markers,
overexpressed_genes=overexpressed_genes)Heatmaps
tested_clusts <- sort(c(intersect_clusters, rescued_clusters))
separation <- c(setNames(rep('rescued', length(rescued_cbs)), rescued_cbs),
setNames(rep('real', length(intersect_clusters)), names(intersect_clusters)))
umis_per_cb_subset <- log10(Matrix::colSums(r_cm_rescued[, names(tested_clusts)]))
tested_clusts <- tested_clusts[order(tested_clusts, -umis_per_cb_subset)]
de_genes <- clusters_info$overexpressed_genesplot_df <- ExpressionMatrixToDataFrame(r_rescued$counts[names(tested_clusts), de_genes],
umis_per_cb_subset, as.factor(tested_clusts),
filtration.type=separation)
plot_df <- plot_df %>% filter(UmisPerCb < 2.83)
plot_dfs <- split(plot_df, plot_df$FiltrationType)ggs <- lapply(plot_dfs, HeatmapAnnotGG, umi.per.cell.limits=range(plot_df$UmisPerCb))
legend_guides <- list(HeatmapLegendGuide('Expression'),
HeatmapLegendGuide('Cell type', guide=guide_legend, ncol=3),
HeatmapLegendGuide('log10(#molecules)'))
gg_legends <- mapply(`+`, ggs$real, legend_guides, SIMPLIFY=F) %>%
lapply(`+`, theme(legend.margin=margin(l=4, r=4, unit='pt'))) %>% lapply(get_legend)
ggs$real$heatmap <- ggs$real$heatmap + rremove('xlab') + ylab('Cells')
ggs$rescued$heatmap <- ggs$rescued$heatmap + labs(x = 'Genes', y = 'Cells')
ggs_annot <- ggs %>%
lapply(function(gg) cowplot::plot_grid(
plotlist=lapply(gg, `+`, theme(legend.position="none", plot.margin=margin())),
nrow=1, rel_widths=c(1.5, 0.1, 0.1), align='h'))Figure
gg_left <- cowplot::plot_grid(ggs_annot$real, ggs_annot$rescued, nrow=2, labels=c('A', 'B'))
gg_right <- gg_tsne + theme(plot.margin=margin(l=0.1, unit='in'),
axis.text=element_blank(), axis.ticks=element_blank())
gg_bottom <- cowplot::plot_grid(plotlist=gg_legends[c(1, 3, 2)], ncol=3,
rel_widths=c(1, 1, 2.2))
gg_fig <- cowplot::plot_grid(gg_left, gg_right, labels=c('', 'C'), ncol=2) %>%
cowplot::plot_grid(gg_bottom, nrow=2, rel_heights=c(1, 0.25), align='v') +
theme(plot.margin=margin(1, 1, 1, 1))gg_fig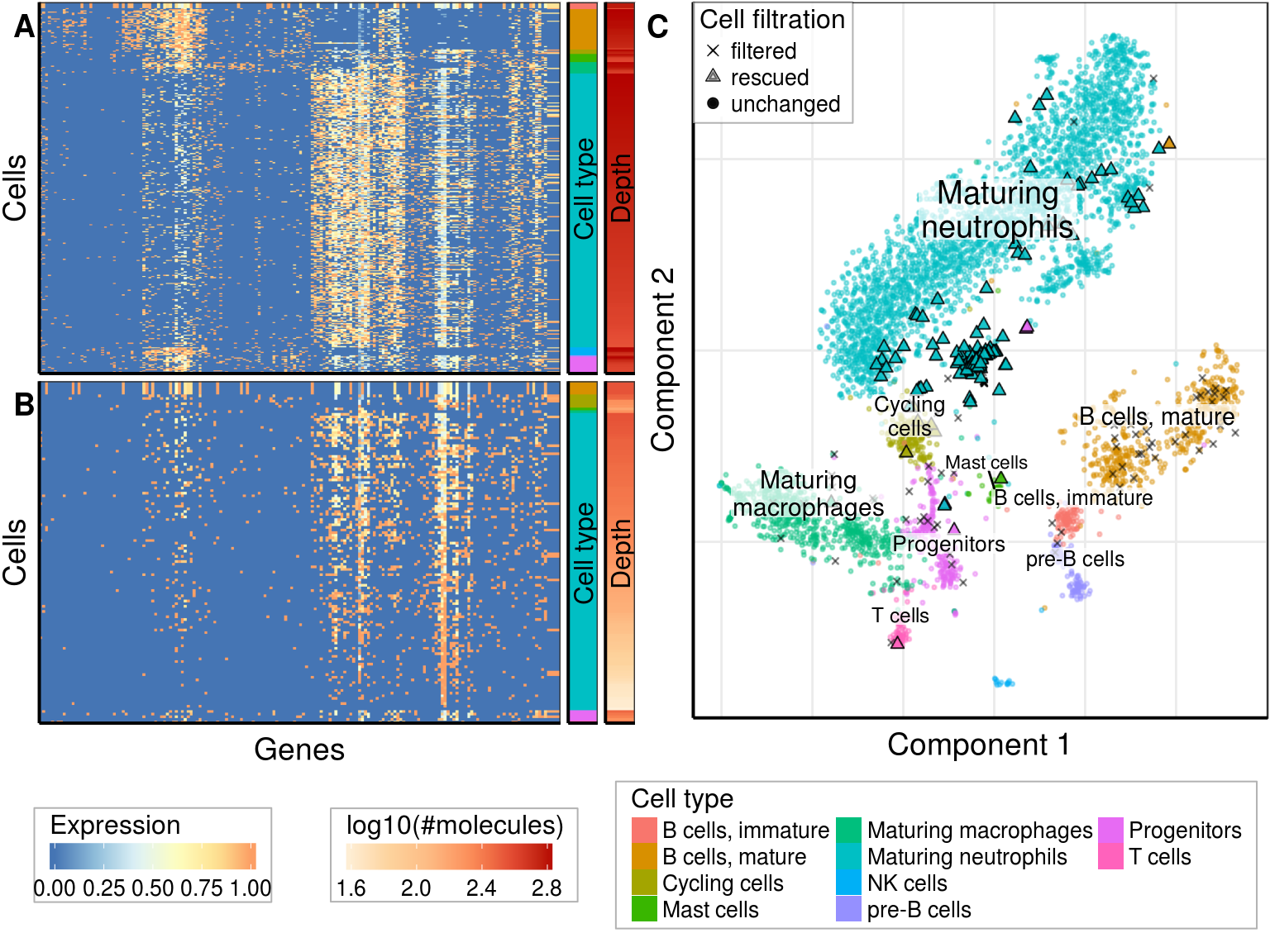
ggsave(paste0(kOutputFolder, 'figures/fig_bmc_lq_cells.pdf'), gg_fig, width=8, height=6)Session information
| value | |
|---|---|
| version | R version 3.4.1 (2017-06-30) |
| os | Ubuntu 14.04.5 LTS |
| system | x86_64, linux-gnu |
| ui | X11 |
| language | (EN) |
| collate | en_US.UTF-8 |
| tz | America/New_York |
| date | 2018-05-16 |
| package | loadedversion | date | source | |
|---|---|---|---|---|
| 1 | acepack | 1.4.1 | 2016-10-29 | CRAN (R 3.4.1) |
| 2 | AnnotationDbi | 1.32.3 | 2016-01-28 | Bioconductor |
| 3 | ape | 5.0 | 2017-10-30 | CRAN (R 3.4.1) |
| 4 | assertthat | 0.2.0 | 2017-04-11 | CRAN (R 3.4.1) |
| 5 | backports | 1.1.2 | 2017-12-13 | CRAN (R 3.4.1) |
| 7 | base64enc | 0.1-3 | 2015-07-28 | cran (@0.1-3) |
| 8 | bindr | 0.1 | 2016-11-13 | CRAN (R 3.4.1) |
| 9 | bindrcpp | 0.2 | 2017-06-17 | CRAN (R 3.4.1) |
| 10 | Biobase | 2.30.0 | 2016-01-28 | Bioconductor |
| 11 | BiocGenerics | 0.16.1 | 2016-01-28 | Bioconductor |
| 12 | bit | 1.1-12 | 2014-04-09 | CRAN (R 3.4.1) |
| 13 | bit64 | 0.9-7 | 2017-05-08 | CRAN (R 3.4.1) |
| 14 | bitops | 1.0-6 | 2013-08-17 | CRAN (R 3.4.1) |
| 15 | blob | 1.1.0 | 2017-06-17 | CRAN (R 3.4.1) |
| 16 | brew | 1.0-6 | 2011-04-13 | CRAN (R 3.4.1) |
| 17 | broom | 0.4.3 | 2017-11-20 | CRAN (R 3.4.1) |
| 18 | Cairo | 1.5-9 | 2015-09-26 | CRAN (R 3.4.1) |
| 19 | caret | 6.0-78 | 2017-12-10 | CRAN (R 3.4.1) |
| 20 | caTools | 1.17.1 | 2014-09-10 | CRAN (R 3.4.1) |
| 21 | checkmate | 1.8.5 | 2017-10-24 | CRAN (R 3.4.1) |
| 22 | class | 7.3-14 | 2015-08-30 | CRAN (R 3.4.0) |
| 23 | clisymbols | 1.2.0 | 2017-05-21 | CRAN (R 3.4.1) |
| 24 | cluster | 2.0.6 | 2017-03-16 | CRAN (R 3.4.0) |
| 25 | codetools | 0.2-15 | 2016-10-05 | CRAN (R 3.4.1) |
| 26 | colorspace | 1.3-2 | 2016-12-14 | CRAN (R 3.4.1) |
| 28 | cowplot | 0.9.2 | 2017-12-17 | CRAN (R 3.4.1) |
| 29 | CVST | 0.2-1 | 2013-12-10 | CRAN (R 3.4.1) |
| 30 | data.table | 1.10.4-3 | 2017-10-27 | CRAN (R 3.4.1) |
| 32 | DBI | 0.7 | 2017-06-18 | CRAN (R 3.4.1) |
| 33 | ddalpha | 1.3.1 | 2017-09-27 | CRAN (R 3.4.1) |
| 34 | dendsort | 0.3.3 | 2015-12-14 | cran (@0.3.3) |
| 35 | DEoptimR | 1.0-8 | 2016-11-19 | CRAN (R 3.4.1) |
| 36 | diffusionMap | 1.1-0 | 2014-02-20 | CRAN (R 3.4.1) |
| 37 | digest | 0.6.15 | 2018-01-28 | cran (@0.6.15) |
| 38 | dimRed | 0.1.0 | 2017-05-04 | CRAN (R 3.4.1) |
| 39 | diptest | 0.75-7 | 2016-12-05 | CRAN (R 3.4.1) |
| 40 | doParallel | 1.0.11 | 2017-09-28 | CRAN (R 3.4.1) |
| 41 | dplyr | 0.7.4 | 2017-09-28 | CRAN (R 3.4.1) |
| 42 | dropEstAnalysis | 0.6.0 | 2018-05-16 | local (VPetukhov/dropEstAnalysis@NA) |
| 43 | dropestr | 0.7.7 | 2018-03-17 | local (@0.7.7) |
| 44 | DRR | 0.0.2 | 2016-09-15 | CRAN (R 3.4.1) |
| 45 | dtw | 1.18-1 | 2015-09-01 | CRAN (R 3.4.1) |
| 46 | evaluate | 0.10.1 | 2017-06-24 | CRAN (R 3.4.1) |
| 47 | flexmix | 2.3-14 | 2017-04-28 | CRAN (R 3.4.1) |
| 48 | FNN | 1.1 | 2013-07-31 | CRAN (R 3.4.1) |
| 49 | foreach | 1.4.4 | 2017-12-12 | CRAN (R 3.4.1) |
| 50 | foreign | 0.8-69 | 2017-06-21 | CRAN (R 3.4.0) |
| 51 | Formula | 1.2-2 | 2017-07-10 | CRAN (R 3.4.1) |
| 52 | fpc | 2.1-10 | 2015-08-14 | CRAN (R 3.4.1) |
| 53 | gdata | 2.18.0 | 2017-06-06 | CRAN (R 3.4.1) |
| 54 | ggjoy | 0.4.0 | 2017-09-15 | CRAN (R 3.4.1) |
| 55 | ggplot2 | 2.2.1 | 2016-12-30 | CRAN (R 3.4.1) |
| 56 | ggpubr | 0.1.6 | 2017-11-14 | CRAN (R 3.4.1) |
| 57 | ggrastr | 0.1.5 | 2017-12-28 | Github (VPetukhov/ggrastr@cc56b45) |
| 58 | ggrepel | 0.7.0 | 2017-09-29 | CRAN (R 3.4.1) |
| 59 | ggridges | 0.4.1 | 2017-09-15 | CRAN (R 3.4.1) |
| 60 | git2r | 0.21.0 | 2018-01-04 | cran (@0.21.0) |
| 61 | glue | 1.2.0 | 2017-10-29 | CRAN (R 3.4.1) |
| 62 | GO.db | 3.2.2 | 2017-11-12 | Bioconductor |
| 63 | gower | 0.1.2 | 2017-02-23 | CRAN (R 3.4.1) |
| 64 | gplots | 3.0.1 | 2016-03-30 | CRAN (R 3.4.1) |
| 68 | gridBase | 0.4-7 | 2014-02-24 | CRAN (R 3.4.1) |
| 69 | gridExtra | 2.3 | 2017-09-09 | CRAN (R 3.4.1) |
| 70 | gtable | 0.2.0 | 2016-02-26 | CRAN (R 3.4.1) |
| 71 | gtools | 3.5.0 | 2015-05-29 | CRAN (R 3.4.1) |
| 72 | highr | 0.6 | 2016-05-09 | CRAN (R 3.4.1) |
| 73 | Hmisc | 4.0-3 | 2017-05-02 | CRAN (R 3.4.1) |
| 74 | htmlTable | 1.11.0 | 2017-12-01 | CRAN (R 3.4.1) |
| 75 | htmltools | 0.3.6 | 2017-04-28 | CRAN (R 3.4.1) |
| 76 | htmlwidgets | 1.0 | 2018-01-20 | cran (@1.0) |
| 77 | ica | 1.0-1 | 2015-08-25 | CRAN (R 3.4.1) |
| 78 | igraph | 1.2.1 | 2018-03-10 | cran (@1.2.1) |
| 79 | ipred | 0.9-6 | 2017-03-01 | CRAN (R 3.4.1) |
| 80 | IRanges | 2.4.8 | 2016-09-15 | Bioconductor |
| 81 | irlba | 2.3.2 | 2018-01-11 | cran (@2.3.2) |
| 82 | iterators | 1.0.9 | 2017-12-12 | CRAN (R 3.4.1) |
| 83 | kernlab | 0.9-25 | 2016-10-03 | CRAN (R 3.4.1) |
| 84 | KernSmooth | 2.23-15 | 2015-06-29 | CRAN (R 3.4.0) |
| 85 | knitr | 1.20 | 2018-02-20 | cran (@1.20) |
| 86 | ks | 1.11.0 | 2018-01-16 | local (VPetukhov/ks@NA) |
| 87 | labeling | 0.3 | 2014-08-23 | CRAN (R 3.4.1) |
| 88 | lars | 1.2 | 2013-04-24 | CRAN (R 3.4.1) |
| 89 | lattice | 0.20-35 | 2017-03-25 | CRAN (R 3.4.1) |
| 90 | latticeExtra | 0.6-28 | 2016-02-09 | CRAN (R 3.4.1) |
| 91 | lava | 1.5.1 | 2017-09-27 | CRAN (R 3.4.1) |
| 92 | lazyeval | 0.2.1 | 2017-10-29 | CRAN (R 3.4.1) |
| 93 | lubridate | 1.7.1 | 2017-11-03 | CRAN (R 3.4.1) |
| 94 | magrittr | 1.5 | 2014-11-22 | CRAN (R 3.4.1) |
| 95 | MASS | 7.3-47 | 2017-04-21 | CRAN (R 3.4.0) |
| 96 | Matrix | 1.2-12 | 2017-11-16 | CRAN (R 3.4.1) |
| 97 | mclust | 5.4 | 2017-11-22 | CRAN (R 3.4.1) |
| 98 | memoise | 1.1.0 | 2017-04-21 | CRAN (R 3.4.1) |
| 100 | mgcv | 1.8-22 | 2017-09-19 | CRAN (R 3.4.1) |
| 101 | mixtools | 1.1.0 | 2017-03-10 | CRAN (R 3.4.1) |
| 102 | mnormt | 1.5-5 | 2016-10-15 | CRAN (R 3.4.1) |
| 103 | ModelMetrics | 1.1.0 | 2016-08-26 | CRAN (R 3.4.1) |
| 104 | modeltools | 0.2-21 | 2013-09-02 | CRAN (R 3.4.1) |
| 105 | munsell | 0.4.3 | 2016-02-13 | CRAN (R 3.4.1) |
| 106 | mvtnorm | 1.0-7 | 2018-01-26 | cran (@1.0-7) |
| 107 | nlme | 3.1-131 | 2017-02-06 | CRAN (R 3.4.0) |
| 108 | NMF | 0.20.6 | 2015-05-26 | CRAN (R 3.4.1) |
| 109 | nnet | 7.3-12 | 2016-02-02 | CRAN (R 3.4.0) |
| 110 | numDeriv | 2016.8-1 | 2016-08-27 | CRAN (R 3.4.1) |
| 111 | pagoda2 | 0.0.0.9002 | 2018-04-08 | local (hms-dbmi/pagoda2@NA) |
| 113 | pbapply | 1.3-3 | 2017-07-04 | CRAN (R 3.4.1) |
| 114 | pcaMethods | 1.60.0 | 2017-11-12 | Bioconductor |
| 115 | pcaPP | 1.9-73 | 2018-01-14 | cran (@1.9-73) |
| 116 | pkgconfig | 2.0.1 | 2017-03-21 | CRAN (R 3.4.1) |
| 117 | pkgmaker | 0.22 | 2014-05-14 | CRAN (R 3.4.1) |
| 118 | plyr | 1.8.4 | 2016-06-08 | CRAN (R 3.4.1) |
| 119 | prabclus | 2.2-6 | 2015-01-14 | CRAN (R 3.4.1) |
| 120 | prodlim | 1.6.1 | 2017-03-06 | CRAN (R 3.4.1) |
| 121 | proxy | 0.4-20 | 2017-12-12 | CRAN (R 3.4.1) |
| 122 | psych | 1.7.8 | 2017-09-09 | CRAN (R 3.4.1) |
| 123 | purrr | 0.2.4 | 2017-10-18 | CRAN (R 3.4.1) |
| 124 | R.methodsS3 | 1.7.1 | 2016-02-16 | CRAN (R 3.4.1) |
| 125 | R.oo | 1.21.0 | 2016-11-01 | CRAN (R 3.4.1) |
| 126 | R.utils | 2.6.0 | 2017-11-05 | CRAN (R 3.4.1) |
| 127 | R6 | 2.2.2 | 2017-06-17 | CRAN (R 3.4.1) |
| 128 | ranger | 0.8.0 | 2017-06-20 | CRAN (R 3.4.1) |
| 129 | RColorBrewer | 1.1-2 | 2014-12-07 | CRAN (R 3.4.1) |
| 130 | Rcpp | 0.12.16 | 2018-03-13 | cran (@0.12.16) |
| 131 | RcppRoll | 0.2.2 | 2015-04-05 | CRAN (R 3.4.1) |
| 132 | recipes | 0.1.1 | 2017-11-20 | CRAN (R 3.4.1) |
| 133 | registry | 0.5 | 2017-12-03 | CRAN (R 3.4.1) |
| 134 | reshape2 | 1.4.3 | 2017-12-11 | CRAN (R 3.4.1) |
| 135 | rjson | 0.2.15 | 2014-11-03 | CRAN (R 3.4.1) |
| 136 | rlang | 0.1.4 | 2017-11-05 | CRAN (R 3.4.1) |
| 137 | rmarkdown | 1.9 | 2018-03-01 | CRAN (R 3.4.1) |
| 138 | rngtools | 1.2.4 | 2014-03-06 | CRAN (R 3.4.1) |
| 139 | robustbase | 0.92-8 | 2017-11-01 | CRAN (R 3.4.1) |
| 140 | ROCR | 1.0-7 | 2015-03-26 | CRAN (R 3.4.1) |
| 141 | Rook | 1.1-1 | 2014-10-20 | CRAN (R 3.4.1) |
| 142 | rpart | 4.1-11 | 2017-04-21 | CRAN (R 3.4.0) |
| 143 | rprojroot | 1.3-2 | 2018-01-03 | cran (@1.3-2) |
| 144 | RSQLite | 2.0 | 2017-06-19 | CRAN (R 3.4.1) |
| 145 | rstudioapi | 0.7 | 2017-09-07 | CRAN (R 3.4.1) |
| 146 | Rtsne | 0.14 | 2017-11-12 | Github (jkrijthe/Rtsne@1d0f926) |
| 147 | S4Vectors | 0.8.11 | 2016-01-30 | Bioconductor |
| 148 | scales | 0.5.0 | 2017-08-24 | CRAN (R 3.4.1) |
| 149 | scatterplot3d | 0.3-40 | 2017-04-22 | CRAN (R 3.4.1) |
| 150 | SDMTools | 1.1-221 | 2014-08-05 | CRAN (R 3.4.1) |
| 151 | segmented | 0.5-3.0 | 2017-11-30 | CRAN (R 3.4.1) |
| 152 | sessioninfo | 1.0.0 | 2017-06-21 | CRAN (R 3.4.1) |
| 153 | Seurat | 2.1.0 | 2017-10-12 | CRAN (R 3.4.1) |
| 154 | sfsmisc | 1.1-1 | 2017-06-08 | CRAN (R 3.4.1) |
| 155 | sn | 1.5-1 | 2017-11-23 | CRAN (R 3.4.1) |
| 159 | stringi | 1.1.7 | 2018-03-12 | cran (@1.1.7) |
| 160 | stringr | 1.3.0 | 2018-02-19 | cran (@1.3.0) |
| 161 | survival | 2.41-3 | 2017-04-04 | CRAN (R 3.4.0) |
| 162 | tclust | 1.3-1 | 2017-08-24 | CRAN (R 3.4.1) |
| 163 | tibble | 1.3.4 | 2017-08-22 | CRAN (R 3.4.1) |
| 164 | tidyr | 0.7.2 | 2017-10-16 | CRAN (R 3.4.1) |
| 165 | tidyselect | 0.2.3 | 2017-11-06 | CRAN (R 3.4.1) |
| 166 | timeDate | 3042.101 | 2017-11-16 | CRAN (R 3.4.1) |
| 168 | triebeard | 0.3.0 | 2016-08-04 | cran (@0.3.0) |
| 169 | trimcluster | 0.1-2 | 2012-10-29 | CRAN (R 3.4.1) |
| 170 | tsne | 0.1-3 | 2016-07-15 | CRAN (R 3.4.1) |
| 171 | urltools | 1.7.0 | 2018-01-20 | cran (@1.7.0) |
| 173 | VGAM | 1.0-4 | 2017-07-25 | CRAN (R 3.4.1) |
| 174 | withr | 2.1.2 | 2018-03-15 | cran (@2.1.2) |
| 175 | xtable | 1.8-2 | 2016-02-05 | CRAN (R 3.4.1) |
| 176 | yaml | 2.1.18 | 2018-03-08 | cran (@2.1.18) |
This R Markdown site was created with workflowr