Drop-seq human/mouse mixture analysis
Viktor Petukhov
2018-01-23
Source file: notebooks/human_mouse/hm_drop_seq.Rmd
Last updated: 2018-02-04
Code version: 0306e1c
library(ggplot2)
library(ggrastr)
library(dplyr)
library(dropestr)
library(dropEstAnalysis)
library(Matrix)
theme_set(theme_base)Load data
Here bam file was filtered by realigning it with kallisto 0.43 separately on mouse and human genome. Only reads, which were aligned only on one of them were used in dropEst.
# holder <- readRDS('../../data/dropest/dropseq/thousand/est_2018_01_26_tophat/thousand.rds')
# saveRDS(holder$cm_raw, '../../data/dropest/dropseq/thousand/est_2018_01_26_tophat/cm.rds')
cm <- readRDS('../../data/dropest/dropseq/thousand/est_2018_01_26_tophat/cm.rds')
kPlotDir <- '../../output/figures/'cell_number <- 1100
gene_species <- ifelse(substr(rownames(cm), 1, 2) == "HU", 'Human', 'Mouse') %>%
as.factor()
umi_by_species <- lapply(levels(gene_species), function(l) cm[gene_species == l,] %>%
Matrix::colSums()) %>% as.data.frame() %>%
`colnames<-`(levels(gene_species)) %>% tibble::rownames_to_column('CB') %>%
as_tibble() %>%
mutate(Total = Human + Mouse, Organism=ifelse(Human > Mouse, "Human", "Mouse"),
IsReal=rank(Total) >= length(Total) - cell_number) %>%
filter(Total > 20)
mit_genes <- rownames(cm)[grep("_MT:.+", rownames(cm))]
umi_by_species$MitochondrionFraction <- GetGenesetFraction(cm, mit_genes)[umi_by_species$CB]
umi_by_species$Type <- ifelse(umi_by_species$IsReal, umi_by_species$Organism, "Background")
umi_by_species$Type[umi_by_species$Mouse > 5e3 & umi_by_species$Human > 5e3] <- 'Dublets'Common view
gg_template <- ggplot(umi_by_species, aes(x=Mouse, y=Human)) +
geom_abline(aes(slope=1, intercept=0), linetype='dashed', alpha=0.5) +
scale_x_log10(limits=c(1, 2e5), name="#Mouse molecules") +
scale_y_log10(name="#Human molecules") + annotation_logticks() +
theme_pdf(legend.pos=c(0.97, 0.05)) + theme(legend.margin=margin(l=3, r=3, unit="pt"))
gg_left <- gg_template +
geom_point(aes(color=IsReal), size=0.1, alpha=0.15) +
guides(color=guide_legend(override.aes=list(size=1.5, alpha=1)))
gg_right <- gg_template +
geom_point(aes(color=MitochondrionFraction), size=0.1, alpha=0.15) +
scale_color_gradientn(colours=c("#1200ba", "#347fff", "#cc4000", "#ff3333"),
values=scales::rescale(c(0, 0.1, 0.3, 0.8))) +
guides(color=guide_colorbar(direction="horizontal", title.position="top",
title="Mitochondrial\nfraction",
barwidth=unit(1.0, units="in")))
cowplot::plot_grid(gg_left, gg_right)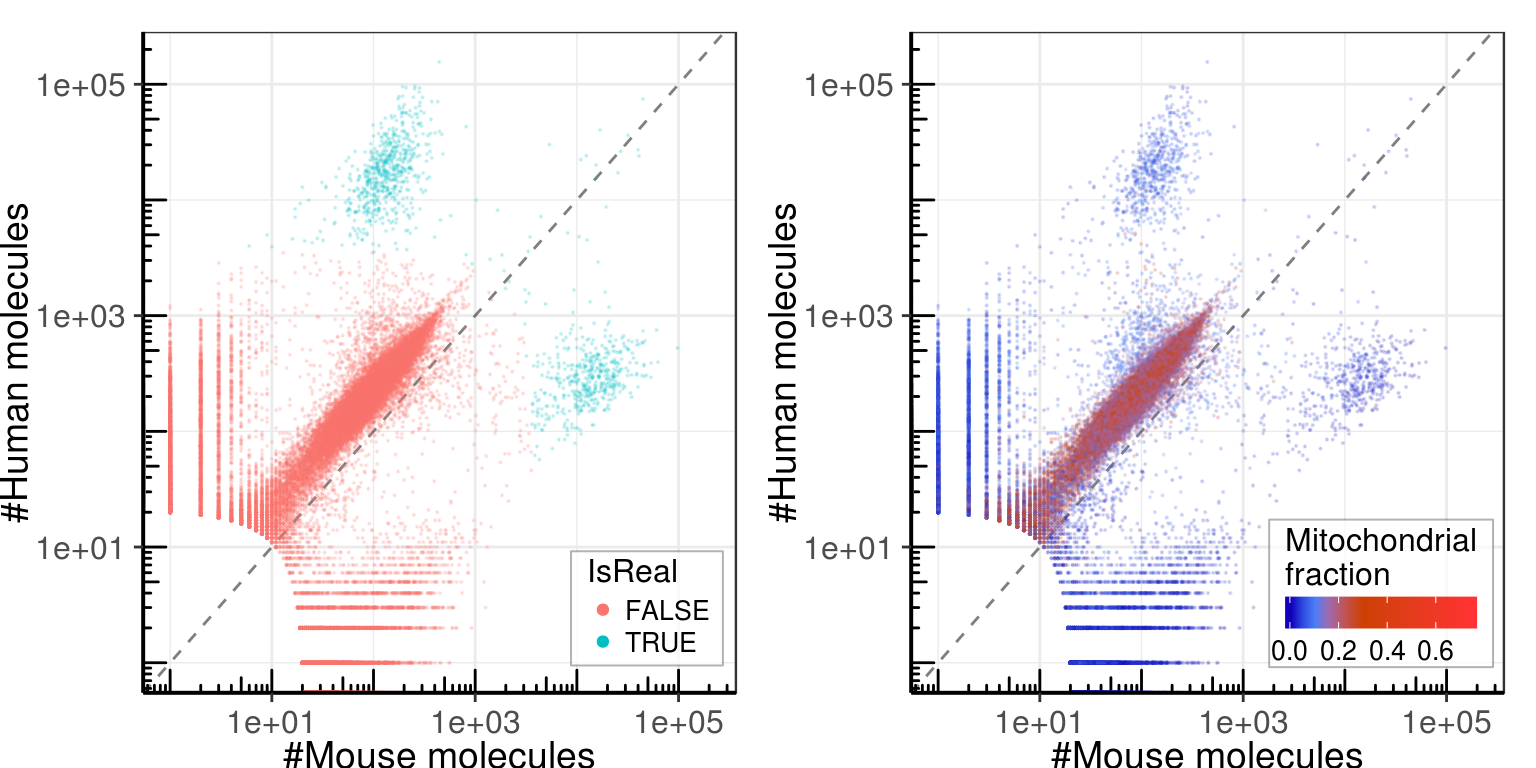
ggplot(umi_by_species) +
geom_point(aes(x=Total, y=pmin(Human, Mouse) / Total, color=Organism), size=0.1,
alpha=0.1) +
scale_x_log10(name='#Real UMIs', limits=c(10, 2e5)) + annotation_logticks() +
ylab('Fraction of mixed UMIs') +
guides(color=guide_legend(override.aes=list(size=1.5, alpha=1))) +
theme_pdf(legend.pos=c(1, 1))
Check for constant background
Background cells have constant fraction of mouse and human reads. Though this fraction is not determined by the corresponding ratio in real cells:
mouse_frac <- umi_by_species %>% filter(IsReal) %>%
summarise(Mouse=sum(Mouse[Organism == 'Mouse']), Human=sum(Human[Organism == 'Human']),
MF=Mouse / (Mouse + Human)) %>% .$MF
ggplot(umi_by_species) +
geom_histogram(aes(x=Mouse / Total, y=..density.., fill=IsReal), binwidth=0.005, position="identity") +
geom_vline(xintercept=mouse_frac) +
xlab("Fraction of mouse reads") +
theme_pdf(legend.pos=c(1, 1))
Distribution of total number of molecules by background cells:
gg <- ggplot(umi_by_species %>% filter(!IsReal)) +
geom_histogram(aes(x=Total), bins=100) +
scale_x_continuous(limits=c(0, 1000), expand=c(0, 0), name="Total #UMIs") +
scale_y_continuous(limits=c(0,4500), expand=c(0, 0), name="#Cells") +
theme_pdf()
gg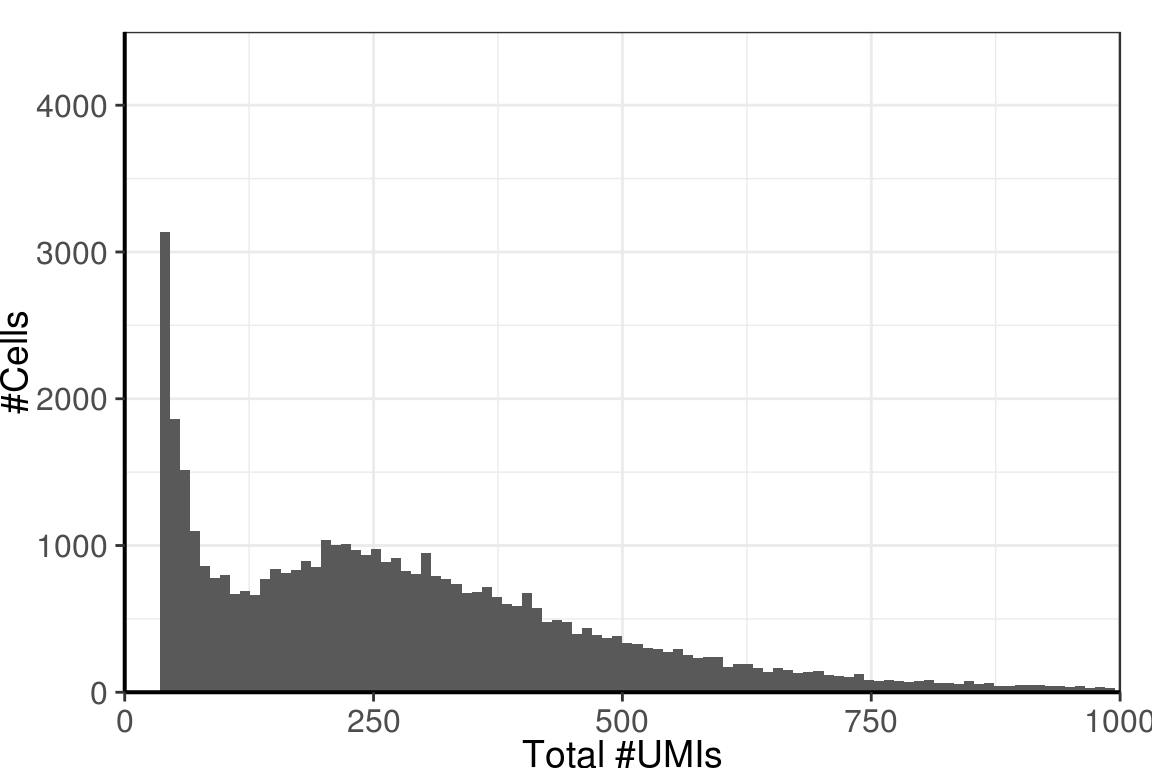
Though, it looks better on logscale:
gg +
scale_x_log10(limits=c(19, 3000), expand=c(0, 0), name="Total #UMIs") +
annotation_logticks(sides="b")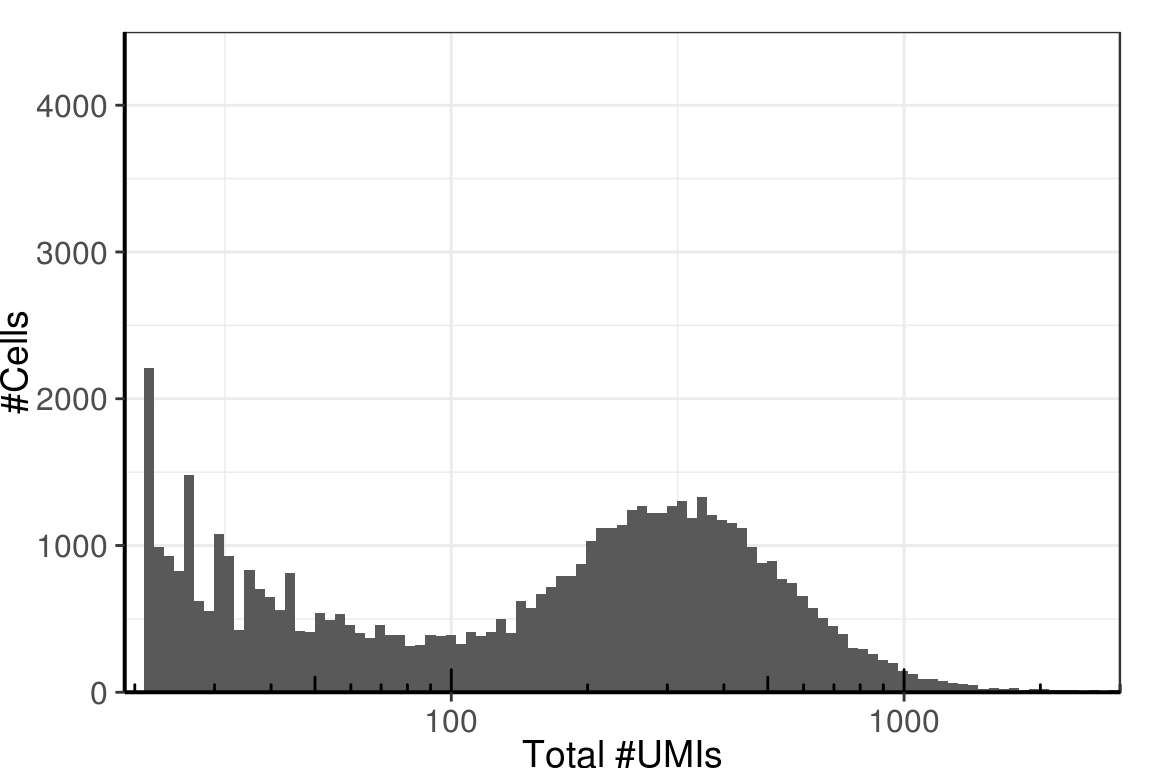
Figure
arrows_df <- umi_by_species %>% group_by(Type) %>%
summarise(MouseEnd=median(Mouse), HumanEnd=median(Human)) %>%
mutate(Mouse=c(1e1, 6e4, 2e1, 6e4),
Human=c(1e3, 5e3, 7e4, 7e1))
fig_width <- 3.7
fig_height <- 4.4
gg_fig <- gg_template +
geom_point_rast(aes(color=MitochondrionFraction), size=0.2, alpha=0.15,
width=fig_width, height=fig_height, dpi=200) +
scale_color_gradientn(colours=c("#1200ba", "#347fff", "#cc4000", "#ff3333"),
values=scales::rescale(c(0, 0.1, 0.3, 0.8)),
breaks=seq(0, 1.0, 0.2)) +
guides(color=guide_colorbar(direction="horizontal", title.position="top",
title="Mitochondrial\nfraction",
barwidth=unit(1.0, units="in"))) +
stat_ellipse(aes(group=Type), level=0.99) +
geom_segment(aes(xend=MouseEnd, yend=HumanEnd, group=Type), data=arrows_df,
arrow=arrow(length = unit(0.03, "npc"))) +
geom_label(aes(label=Type), data=arrows_df, fill=alpha('white', 1)) +
theme(plot.margin=margin(1, 1, 1, 1))
try(invisible(dev.off()), silent=T)
ggsave(paste0(kPlotDir, 'fig_human_mouse.pdf'), gg_fig,
width=fig_width, height=fig_height)gg_fig
Session information
| value | |
|---|---|
| version | R version 3.4.1 (2017-06-30) |
| os | Ubuntu 14.04.5 LTS |
| system | x86_64, linux-gnu |
| ui | X11 |
| language | (EN) |
| collate | en_US.UTF-8 |
| tz | America/New_York |
| date | 2018-02-04 |
| package | loadedversion | date | source | |
|---|---|---|---|---|
| 1 | assertthat | 0.2.0 | 2017-04-11 | CRAN (R 3.4.1) |
| 2 | backports | 1.1.2 | 2017-12-13 | CRAN (R 3.4.1) |
| 4 | bindr | 0.1 | 2016-11-13 | CRAN (R 3.4.1) |
| 5 | bindrcpp | 0.2 | 2017-06-17 | CRAN (R 3.4.1) |
| 6 | Cairo | 1.5-9 | 2015-09-26 | CRAN (R 3.4.1) |
| 7 | clisymbols | 1.2.0 | 2017-05-21 | CRAN (R 3.4.1) |
| 8 | colorspace | 1.3-2 | 2016-12-14 | CRAN (R 3.4.1) |
| 10 | cowplot | 0.9.2 | 2017-12-17 | CRAN (R 3.4.1) |
| 12 | digest | 0.6.14 | 2018-01-14 | cran (@0.6.14) |
| 13 | dplyr | 0.7.4 | 2017-09-28 | CRAN (R 3.4.1) |
| 14 | dropEstAnalysis | 0.6.0 | 2018-02-01 | local (VPetukhov/dropEstAnalysis@NA) |
| 15 | dropestr | 0.7.5 | 2018-01-31 | local (@0.7.5) |
| 16 | evaluate | 0.10.1 | 2017-06-24 | CRAN (R 3.4.1) |
| 17 | ggplot2 | 2.2.1 | 2016-12-30 | CRAN (R 3.4.1) |
| 18 | ggrastr | 0.1.5 | 2017-12-28 | Github (VPetukhov/ggrastr@cc56b45) |
| 19 | git2r | 0.21.0 | 2018-01-04 | cran (@0.21.0) |
| 20 | glue | 1.2.0 | 2017-10-29 | CRAN (R 3.4.1) |
| 24 | gtable | 0.2.0 | 2016-02-26 | CRAN (R 3.4.1) |
| 25 | highr | 0.6 | 2016-05-09 | CRAN (R 3.4.1) |
| 26 | htmltools | 0.3.6 | 2017-04-28 | CRAN (R 3.4.1) |
| 27 | knitr | 1.18 | 2017-12-27 | cran (@1.18) |
| 28 | labeling | 0.3 | 2014-08-23 | CRAN (R 3.4.1) |
| 29 | lattice | 0.20-35 | 2017-03-25 | CRAN (R 3.4.1) |
| 30 | lazyeval | 0.2.1 | 2017-10-29 | CRAN (R 3.4.1) |
| 31 | magrittr | 1.5 | 2014-11-22 | CRAN (R 3.4.1) |
| 32 | MASS | 7.3-47 | 2017-04-21 | CRAN (R 3.4.0) |
| 33 | Matrix | 1.2-12 | 2017-11-16 | CRAN (R 3.4.1) |
| 35 | munsell | 0.4.3 | 2016-02-13 | CRAN (R 3.4.1) |
| 36 | pkgconfig | 2.0.1 | 2017-03-21 | CRAN (R 3.4.1) |
| 37 | plyr | 1.8.4 | 2016-06-08 | CRAN (R 3.4.1) |
| 38 | R6 | 2.2.2 | 2017-06-17 | CRAN (R 3.4.1) |
| 39 | Rcpp | 0.12.15 | 2018-01-20 | cran (@0.12.15) |
| 40 | rlang | 0.1.4 | 2017-11-05 | CRAN (R 3.4.1) |
| 41 | rmarkdown | 1.8 | 2017-11-17 | CRAN (R 3.4.1) |
| 42 | rprojroot | 1.3-2 | 2018-01-03 | cran (@1.3-2) |
| 43 | scales | 0.5.0 | 2017-08-24 | CRAN (R 3.4.1) |
| 44 | sessioninfo | 1.0.0 | 2017-06-21 | CRAN (R 3.4.1) |
| 46 | stringi | 1.1.6 | 2017-11-17 | CRAN (R 3.4.1) |
| 47 | stringr | 1.2.0 | 2017-02-18 | CRAN (R 3.4.1) |
| 48 | tibble | 1.3.4 | 2017-08-22 | CRAN (R 3.4.1) |
| 51 | withr | 2.1.1 | 2017-12-19 | cran (@2.1.1) |
| 52 | yaml | 2.1.16 | 2017-12-12 | CRAN (R 3.4.1) |
This R Markdown site was created with workflowr